The METLIN small molecule dataset for machine learning-based retention time prediction.


The Metsys Lab is part of the Omics Sciences Unit at The Technology Centre of Catalonia (EURECAT), and the Department of Electrical, Electronic and Control Engineering at the University Rovira i Virgili (URV). We combine bioinformatics and analytical chemistry to develop computational tools applied to mass spectrometry-based metabolomics.
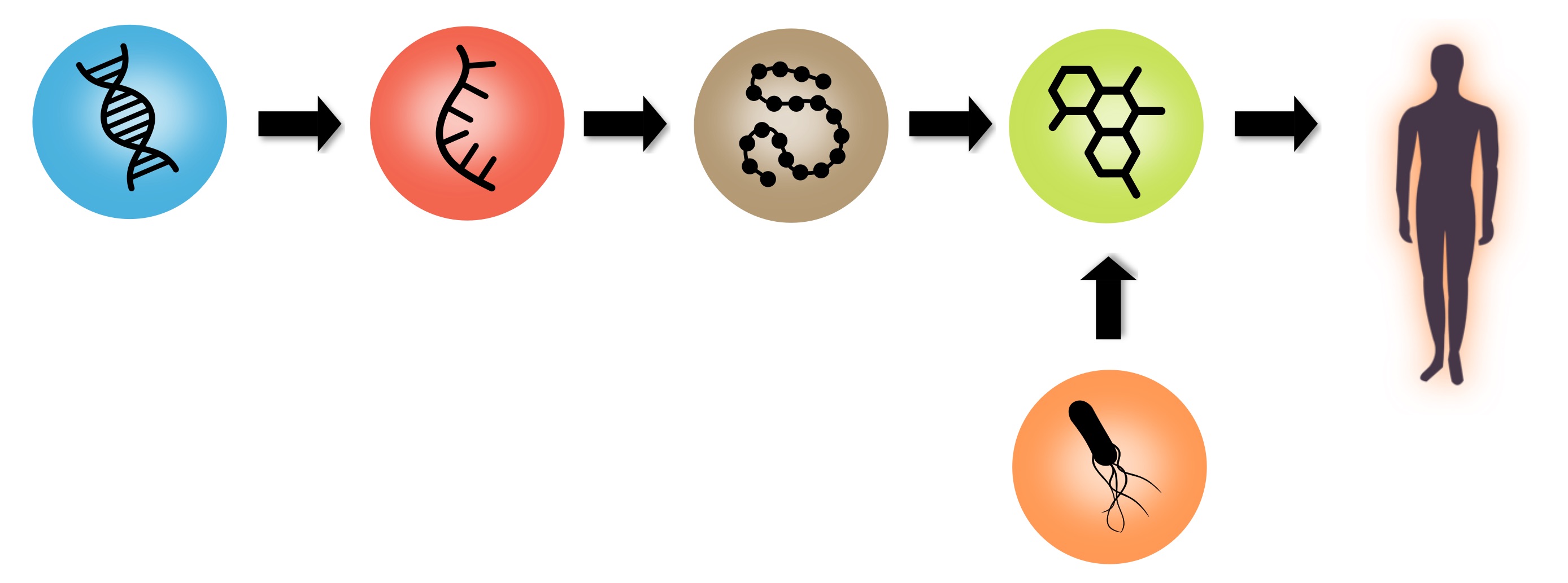
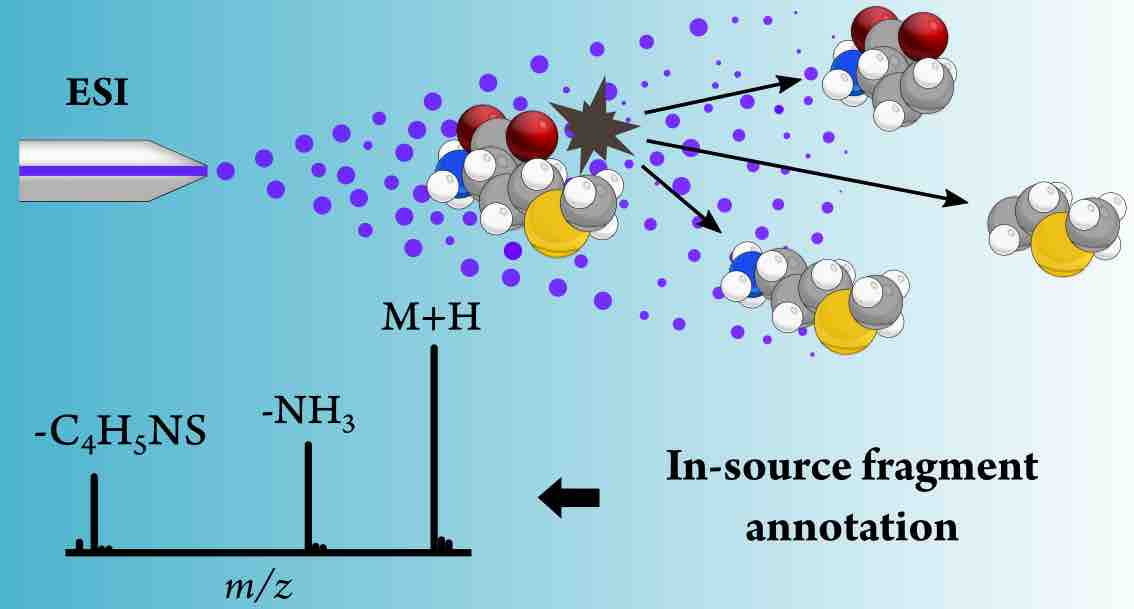
Metabolites are the downstream product of cellular biochemistry. At the same time, metabolites are the central hub in microbiome-host interactions. The identification (annotation) of metabolites in biomedical experiments is the cornerstone for gaining new insights into cell biology, physiology and medicine. Mass spectrometry-based metabolomics is a widely used technique that allows measuring metabolites in biofluids and tissues from living organisms. At the Metsys lab we combine signal processing, machine learning, statistical and analytical chemistry techniques to develop bioinformatics tools and resources for mass spectrometry-based metabolomics data analysis and metabolite identification.
Our lab is supported by:

 |
 |
 |
|---|---|---|
SIGNAL PROCESSING |
MACHINE LEARNING |
ANALYTICAL CHEMISTRY |
| To process mass spectrometry signals and leverage their multivariate nature to enhance data processing performance. | To predict chemical properties or analyze complex molecular interactions. | To increase the algorithm's performance by designing specific analytical solutions and methods. |
 |
 |
|---|---|
SYSTEMS BIOLOGY |
STATISTICS |
| To understand complex biological interactions through multi-omics data integration. | To make our methods robust and assess the accuracy of our algorithms and findings. |